require(ospsuite.reportingengine)
#> Loading required package: ospsuite.reportingengine
#> Loading required package: tlf
#> Loading required package: ospsuiteThis vignette focuses on time profiles and residuals plots (also called goodness of fit plots) generated using either mean model or population workflows.
Overview
The time profiles and residuals task first aims at generating time profile plots of specified simulation paths. The task can account for observed data, in which case residuals are calculated and plotted. For population workflows, aggregation functions are used, by default mean, median, 5th and 95th percentiles are computed and plotted in the time profiles.
Inputs
Results obtained from the simulate task and stored as
csv files within the subdirectory SimulationResults
from the workflowFolder directory are required in order to
perform the goodness of fit (plotTimeProfilesAndResiduals)
task. As a consequence, if the workflow output folder does not include
the appropriate simulations, the task simulate needs to be
active.
The objects SimulationSet (or
PopulationSimulationSet for population workflows) and
Output define which and how the simulated and, if
available, observed data will be plotted.
Outputs
The time profiles and residuals task generates for each simulation set plots of:
- the simulated time profile
- in linear and logarithmic scale
- if available with data
- for total simulated time range.
Multiple applications
For simulations with multiple applications, up to 6 time profile plots can be generated corresponding to simulated time profiles in linear and logarithmic scale for:
- the total simulated time range
- the time range of the first application
- the time range of the last application
The input applicationRanges from
SimulationSet objects can be used to select which ranges to
include in the evaluation.
Residuals
If observed data are available in the range selected in the output
definition and selected in dataSelection, they will be
added to the time profile plots.
Furthermore, residuals will be calculated according to the residual
scale defined by the Output objects included within the
simulation sets and 6 additional plots will be created figures:
- Predicted vs observed in linear scale
- Predicted vs observed in log scale
- Residuals vs time
- Residuals vs simulated output
- Histogram of residuals
- qq-plot of residuals
The residual scale corresponds to one element from the enum
ResidualScales:
ResidualScales
#> $Linear
#> [1] "Linear"
#>
#> $Logarithmic
#> [1] "Logarithmic"The default uses “Logarithmic” and calculate residuals as:
If the option “Linear” is used instead, the residuals are calculated as:
A histogram and a qq-plot of residuals across all the simulation sets will also be added at the end of the analysis.
For population workflows, residuals are calculated from the aggregation of the Simulated output. By default, is used instead of . It is possible to modify this aggregation function by updating the fields of the object AggregationConfiguration.
Template examples
The following sections will introduce template scripts including goodness of fit tasks as well as their associated reports. Table 1 shows the features tested in each template script.
| Example | Workflow | Simulation Sets | Outputs | Observed Data |
|---|---|---|---|---|
| 1 | Mean model | 1 | 1 | No |
| 2 | Mean model | 1 | 1 | Yes |
| 3 | Mean model | 1 | 1 | Yes |
| 4 | Mean model | 1 | 2 | Yes |
| 5 | Mean model | 2 | 1 | Yes |
| 6 | Population model | 1 | 1 | No |
| 7 | Population model | 1 | 1 | Yes |
Example 1
Example 1 shows one of the simplest workflows including a goodness of fit. In this example, only a simulation time profile is performed since there is no observed data. Furthermore, only one simulation set and one output are requested.
Code
# Get the pkml simulation file: "MiniModel2.pkml"
simulationFile <- system.file("extdata", "MiniModel2.pkml",
package = "ospsuite.reportingengine"
)
# Define the input parameters
outputA <- Output$new(
path = "Organism|A|Concentration in container",
displayName = "Concentration of A",
displayUnit = "nmol/ml"
)
setA <- SimulationSet$new(
simulationSetName = "A",
simulationFile = simulationFile,
outputs = outputA
)
# Create the workflow instance
workflow1 <-
MeanModelWorkflow$new(
simulationSets = setA,
workflowFolder = "Example-1"
)
#> 16/01/2026 - 14:33:48
#> i Info Reporting Engine Information:
#> Date: 16/01/2026 - 14:33:48
#> User Information:
#> Computer Name: runnervmmtnos
#> User: runner
#> Login: unknown
#> System is NOT validated
#> System versions:
#> R version: R version 4.5.2 (2025-10-31)
#> OSP Suite Package version: 12.4.1.9001
#> OSP Reporting Engine version: 2.4.0.9005
#> tlf version: 1.6.2.9001
# Set the workflow tasks to be run
workflow1$activateTasks(c("simulate", "plotTimeProfilesAndResiduals"))
# Run the workflow
workflow1$runWorkflow()
#> 16/01/2026 - 14:33:48
#> i Info Starting run of Mean Model Workflow
#> 16/01/2026 - 14:33:48
#> i Info Starting run of Simulation task
#> 16/01/2026 - 14:33:48
#> i Info Splitting simulations for parallel run: 1 simulations split into 1 subsets
#> 16/01/2026 - 14:33:48
#> i Info Starting run of subset simulations
#> | | | 0% | |======================================================================| 100%
#> 16/01/2026 - 14:33:49
#> i Info Simulation task completed in 0 min
#> 16/01/2026 - 14:33:49
#> i Info Starting run of Plot Time profiles and Residuals task
#> 16/01/2026 - 14:33:49
#> i Info Starting run of Plot Time profiles and Residuals task for A
#> 16/01/2026 - 14:33:51
#> i Info Plot Time profiles and Residuals task completed in 0 min
#> 16/01/2026 - 14:33:51
#> i Info Executing: pandoc --embed-resources --standalone --wrap=none --toc --from=markdown+tex_math_dollars+superscript+subscript+raw_attribute --reference-doc="/home/runner/work/_temp/Library/ospsuite.reportingengine/extdata/reference.docx" --resource-path="Example-1" -t docx -o 'Example-1/Report-word.docx' 'Example-1/Report-word.md'
#>
#> 16/01/2026 - 14:33:51
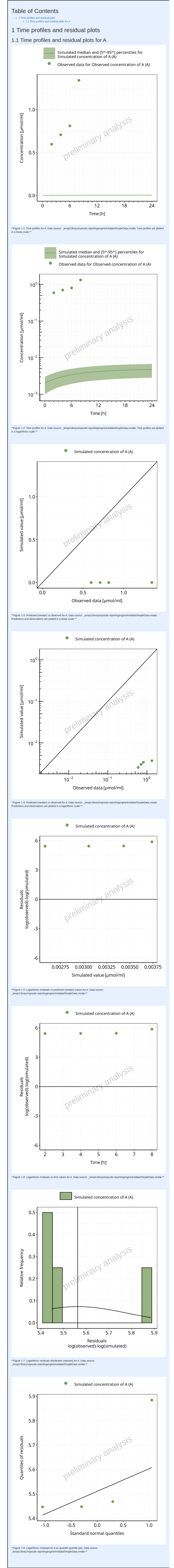
#> i Info Mean Model Workflow completed in 0.1 minThe output report for Example 1 is shown below. There are only 2 figures in the report (linear and log scales) showing the time profile of the output path ‘Organism|A|Concentration in container’ displayed as ‘Concentration of A’.
#> file:////home/runner/work/OSPSuite.ReportingEngine/OSPSuite.ReportingEngine/vignettes/Example-1/Report.html screenshot completedReport
Example 2
Example 2 shows a workflow that used the simulation results performed in Example 1. This example shows 3 new features:
- The presence of observed data compared to the simulation
- The use of results obtained from a previous workflow
- The change of display unit without re-running the
simulatetask
Code
# Get the pkml simulation file: "MiniModel2.pkml"
simulationFile <- system.file("extdata", "MiniModel2.pkml",
package = "ospsuite.reportingengine"
)
# Get the observation file and its dictionary
observationFile <- system.file(
"extdata", "SimpleData.nmdat",
package = "ospsuite.reportingengine"
)
dictionaryFile <- system.file(
"extdata", "tpDictionary.csv",
package = "ospsuite.reportingengine"
)
dataSource <- DataSource$new(
dataFile = observationFile,
metaDataFile = dictionaryFile
)
# Define the input parameters
outputA <- Output$new(
path = "Organism|A|Concentration in container",
displayName = "Simulated concentration of A",
displayUnit = "µmol/ml",
dataSelection = "MDV==0",
dataDisplayName = "Observed concentration of A"
)
setA <- SimulationSet$new(
simulationSetName = "A",
simulationFile = simulationFile,
outputs = outputA,
dataSource = dataSource
)
#> Warning in readLines(file): incomplete final line found on
#> '/home/runner/work/_temp/Library/ospsuite.reportingengine/extdata/tpDictionary.csv'
# Create the workflow instance
workflow2 <-
MeanModelWorkflow$new(
simulationSets = setA,
workflowFolder = "Example-1"
)
#> 16/01/2026 - 14:33:53
#> i Info Reporting Engine Information:
#> Date: 16/01/2026 - 14:33:53
#> User Information:
#> Computer Name: runnervmmtnos
#> User: runner
#> Login: unknown
#> System is NOT validated
#> System versions:
#> R version: R version 4.5.2 (2025-10-31)
#> OSP Suite Package version: 12.4.1.9001
#> OSP Reporting Engine version: 2.4.0.9005
#> tlf version: 1.6.2.9001
# Set the workflow tasks to be run
workflow2$activateTasks("plotTimeProfilesAndResiduals")
workflow2$inactivateTasks("simulate")
# Run the workflow
workflow2$runWorkflow()
#> 16/01/2026 - 14:33:54
#> i Info Starting run of Mean Model Workflow
#> 16/01/2026 - 14:33:54
#> i Info Starting run of Plot Time profiles and Residuals task
#> 16/01/2026 - 14:33:54
#> i Info Starting run of Plot Time profiles and Residuals task for A
#> 16/01/2026 - 14:33:54
#> ! Warning incomplete final line found on '/home/runner/work/_temp/Library/ospsuite.reportingengine/extdata/tpDictionary.csv'
#> 16/01/2026 - 14:34:03
#> i Info Plot Time profiles and Residuals task completed in 0.2 min
#> 16/01/2026 - 14:34:03
#> i Info Executing: pandoc --embed-resources --standalone --wrap=none --toc --from=markdown+tex_math_dollars+superscript+subscript+raw_attribute --reference-doc="/home/runner/work/_temp/Library/ospsuite.reportingengine/extdata/reference.docx" --resource-path="Example-1" -t docx -o 'Example-1/Report-word.docx' 'Example-1/Report-word.md'
#>
#> 16/01/2026 - 14:34:03
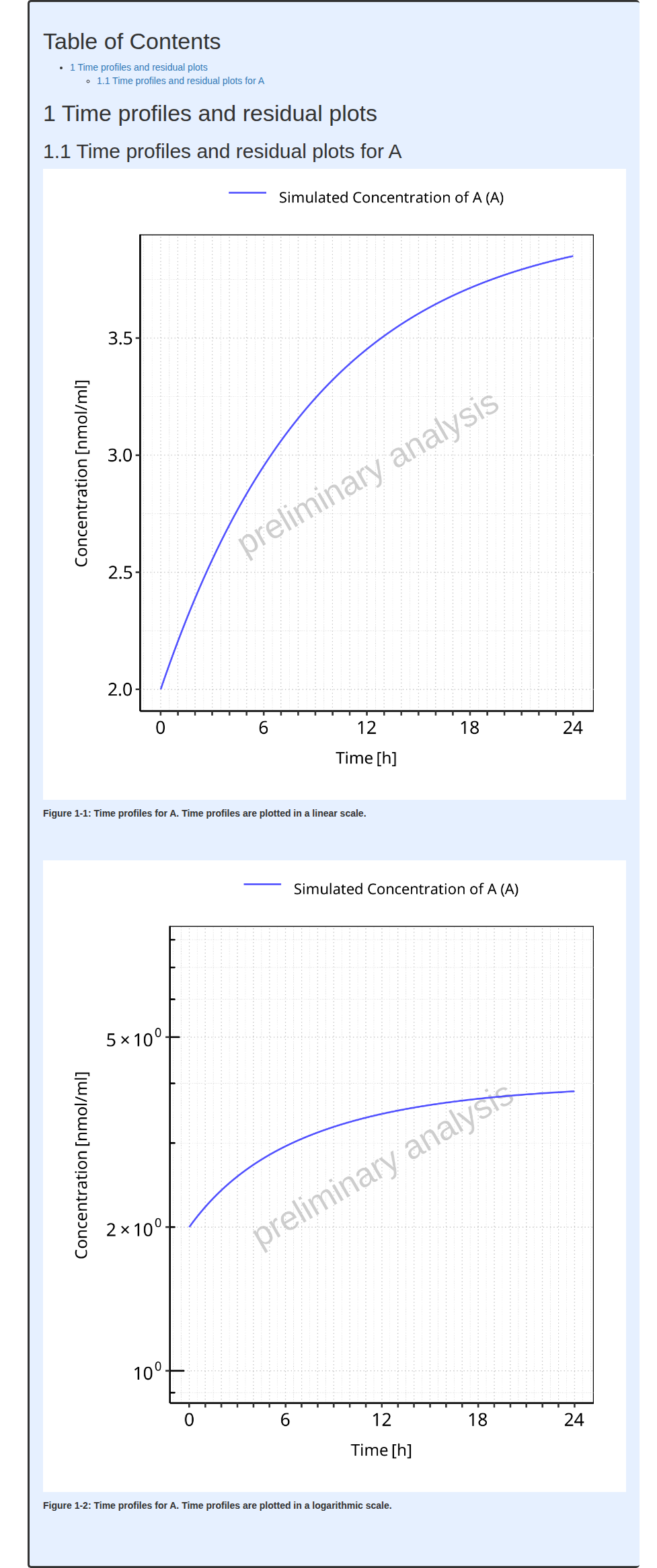
#> i Info Mean Model Workflow completed in 0.2 minIt can be seen from the run results that the task
simulate was not run.
In this example, the content of the observed data file and its dictionary are respectively described Table 2 and 3.
| ID | STUD | SID | TIME | TAD | DV | CMT | MDV | AMT | WGHT | HGHT | SEX | AGE | LOQ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 9999 | 1 | 0 | 0 | 0.00000 | 2 | 1 | 450 | 82 | 160 | 1 | 32 | 0.001 |
| 2 | 9999 | 1 | 2 | 2 | 0.00120 | 1 | 0 | 0 | 82 | 160 | 1 | 32 | 0.001 |
| 3 | 9999 | 1 | 4 | 4 | 0.00142 | 1 | 0 | 0 | 82 | 160 | 1 | 32 | 0.001 |
| 4 | 9999 | 1 | 6 | 6 | 0.00163 | 1 | 0 | 0 | 82 | 160 | 1 | 32 | 0.001 |
| 5 | 9999 | 1 | 8 | 8 | 0.00269 | 1 | 0 | 0 | 82 | 160 | 1 | 32 | 0.001 |
#> Warning in readLines(file): incomplete final line found on
#> '/home/runner/work/_temp/Library/ospsuite.reportingengine/extdata/tpDictionary.csv'| ID | type | datasetColumn | datasetUnit | reportName | pathID | comment |
|---|---|---|---|---|---|---|
| sid | identifier | SID | ||||
| stud | identifier | STUD | ||||
| time | timeprofile | TIME | h | |||
| dv | timeprofile | DV | mg/ml | |||
| tad | timeprofile | TAD | h | |||
| age | covariate | AGE | year(s) | Age | Organism|Age | |
| wght | covariate | WGHT | kg | Body weight | Organism|Weight | |
| hght | covariate | HGHT | cm | Height | Organism|Height | |
| bmi | covariate | BMI | kg/m² | BMI | Organism|BMI | |
| gender | covariate | SEX | SEX | Gender | Make sure male=MALE and female=FEMALE |
The output report for Example 2 is shown below. There are now many
more figures in the report showing not only time profiles in
µmol/ml instead of nmol/ml but also
residuals.
#> file:////home/runner/work/OSPSuite.ReportingEngine/OSPSuite.ReportingEngine/vignettes/Example-1/Report.html screenshot completedReport
Example 3
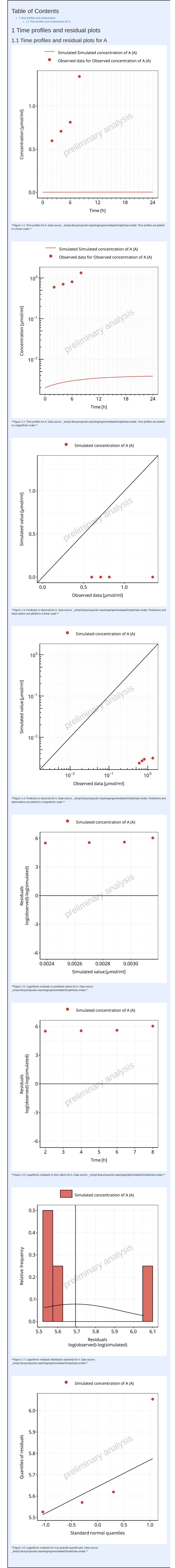
In example 3, the dictionary is changed and indicates that the lower
limit of quantitation (LLOQ) is included in the observed data. Like the
previous example, there is no need to re-run the simulate
task.
Code
# Get the pkml simulation file: "MiniModel2.pkml"
simulationFile <- system.file("extdata", "MiniModel2.pkml",
package = "ospsuite.reportingengine"
)
# Get the observation file and its dictionary
dataSource <- DataSource$new(
dataFile = observationFile,
metaDataFile = system.file(
"extdata", "tpDictionaryLoq.csv",
package = "ospsuite.reportingengine"
)
)
# Define the input parameters
outputA <- Output$new(
path = "Organism|A|Concentration in container",
displayName = "Simulated concentration of A",
displayUnit = "µmol/ml",
dataSelection = "MDV==0",
dataDisplayName = "Observed concentration of A"
)
setA <- SimulationSet$new(
simulationSetName = "A",
simulationFile = simulationFile,
outputs = outputA,
dataSource = dataSource
)
#> Warning in readLines(file): incomplete final line found on
#> '/home/runner/work/_temp/Library/ospsuite.reportingengine/extdata/tpDictionaryLoq.csv'
# Create the workflow instance
workflow3 <-
MeanModelWorkflow$new(
simulationSets = setA,
workflowFolder = "Example-1"
)
#> 16/01/2026 - 14:34:05
#> i Info Reporting Engine Information:
#> Date: 16/01/2026 - 14:34:05
#> User Information:
#> Computer Name: runnervmmtnos
#> User: runner
#> Login: unknown
#> System is NOT validated
#> System versions:
#> R version: R version 4.5.2 (2025-10-31)
#> OSP Suite Package version: 12.4.1.9001
#> OSP Reporting Engine version: 2.4.0.9005
#> tlf version: 1.6.2.9001
# Set the workflow tasks to be run
workflow3$activateTasks("plotTimeProfilesAndResiduals")
workflow3$inactivateTasks("simulate")
# Run the workflow
workflow3$runWorkflow()
#> 16/01/2026 - 14:34:05
#> i Info Starting run of Mean Model Workflow
#> 16/01/2026 - 14:34:05
#> i Info Starting run of Plot Time profiles and Residuals task
#> 16/01/2026 - 14:34:05
#> i Info Starting run of Plot Time profiles and Residuals task for A
#> 16/01/2026 - 14:34:05
#> ! Warning incomplete final line found on '/home/runner/work/_temp/Library/ospsuite.reportingengine/extdata/tpDictionaryLoq.csv'
#> 16/01/2026 - 14:34:14
#> i Info Plot Time profiles and Residuals task completed in 0.2 min
#> 16/01/2026 - 14:34:14
#> i Info Executing: pandoc --embed-resources --standalone --wrap=none --toc --from=markdown+tex_math_dollars+superscript+subscript+raw_attribute --reference-doc="/home/runner/work/_temp/Library/ospsuite.reportingengine/extdata/reference.docx" --resource-path="Example-1" -t docx -o 'Example-1/Report-word.docx' 'Example-1/Report-word.md'
#>
#> 16/01/2026 - 14:34:14
#> i Info Mean Model Workflow completed in 0.2 minThe new dictionary content is described in Table 4.
#> Warning in readLines(file): incomplete final line found on
#> '/home/runner/work/_temp/Library/ospsuite.reportingengine/extdata/tpDictionary.csv'| ID | type | datasetColumn | datasetUnit | reportName | pathID | comment |
|---|---|---|---|---|---|---|
| sid | identifier | SID | ||||
| stud | identifier | STUD | ||||
| time | timeprofile | TIME | h | |||
| dv | timeprofile | DV | mg/ml | |||
| tad | timeprofile | TAD | h | |||
| age | covariate | AGE | year(s) | Age | Organism|Age | |
| wght | covariate | WGHT | kg | Body weight | Organism|Weight | |
| hght | covariate | HGHT | cm | Height | Organism|Height | |
| bmi | covariate | BMI | kg/m² | BMI | Organism|BMI | |
| gender | covariate | SEX | SEX | Gender | Make sure male=MALE and female=FEMALE |
The output report for Example 3 is shown below. The time profiles now indicate the LLOQ.
#> file:////home/runner/work/OSPSuite.ReportingEngine/OSPSuite.ReportingEngine/vignettes/Example-1/Report.html screenshot completedReport
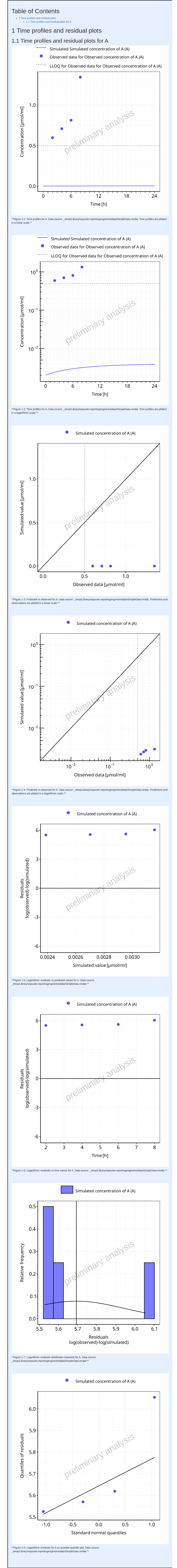
Example 4
In example 4, the simulation set now includes 2 output paths. Output
path Organism|A|Concentration in container has observed
data to be compared to whereas output path
Organism|B|Concentration in container does not. Since
output path Organism|B|Concentration in container was
not requested by the examples 1, 2 and 3, it is not included in the
simulation results. As a consequence, the simulate task
needs to be re-run.
Code
# Get the pkml simulation file: "MiniModel2.pkml"
simulationFile <- system.file("extdata", "MiniModel2.pkml",
package = "ospsuite.reportingengine"
)
# Get the observation file and its dictionary
dataSource <- DataSource$new(
dataFile = observationFile,
metaDataFile = dictionaryFile
)
# Define the input parameters
outputA <- Output$new(
path = "Organism|A|Concentration in container",
displayName = "Simulated concentration of A",
displayUnit = "µmol/ml",
dataSelection = "MDV==0",
dataDisplayName = "Observed concentration of A"
)
outputB <- Output$new(
path = "Organism|B|Concentration in container",
displayName = "Simulated concentration of B",
displayUnit = "µmol/ml"
)
setAB <- SimulationSet$new(
simulationSetName = "A and B",
simulationFile = simulationFile,
outputs = c(outputA, outputB),
dataSource = dataSource
)
#> Warning in readLines(file): incomplete final line found on
#> '/home/runner/work/_temp/Library/ospsuite.reportingengine/extdata/tpDictionary.csv'
# Create the workflow instance
workflow4 <-
MeanModelWorkflow$new(
simulationSets = setAB,
workflowFolder = "Example-4"
)
#> 16/01/2026 - 14:34:16
#> i Info Reporting Engine Information:
#> Date: 16/01/2026 - 14:34:16
#> User Information:
#> Computer Name: runnervmmtnos
#> User: runner
#> Login: unknown
#> System is NOT validated
#> System versions:
#> R version: R version 4.5.2 (2025-10-31)
#> OSP Suite Package version: 12.4.1.9001
#> OSP Reporting Engine version: 2.4.0.9005
#> tlf version: 1.6.2.9001
# Set the workflow tasks to be run
workflow4$activateTasks(c("simulate", "plotTimeProfilesAndResiduals"))
# Run the workflow
workflow4$runWorkflow()
#> 16/01/2026 - 14:34:16
#> i Info Starting run of Mean Model Workflow
#> 16/01/2026 - 14:34:16
#> i Info Starting run of Simulation task
#> 16/01/2026 - 14:34:16
#> i Info Splitting simulations for parallel run: 1 simulations split into 1 subsets
#> 16/01/2026 - 14:34:16
#> i Info Starting run of subset simulations
#> | | | 0% | |======================================================================| 100%
#> 16/01/2026 - 14:34:17
#> i Info Simulation task completed in 0 min
#> 16/01/2026 - 14:34:17
#> i Info Starting run of Plot Time profiles and Residuals task
#> 16/01/2026 - 14:34:17
#> i Info Starting run of Plot Time profiles and Residuals task for A and B
#> 16/01/2026 - 14:34:17
#> ! Warning incomplete final line found on '/home/runner/work/_temp/Library/ospsuite.reportingengine/extdata/tpDictionary.csv'
#> 16/01/2026 - 14:34:28
#> i Info Plot Time profiles and Residuals task completed in 0.2 min
#> 16/01/2026 - 14:34:28
#> i Info Executing: pandoc --embed-resources --standalone --wrap=none --toc --from=markdown+tex_math_dollars+superscript+subscript+raw_attribute --reference-doc="/home/runner/work/_temp/Library/ospsuite.reportingengine/extdata/reference.docx" --resource-path="Example-4" -t docx -o 'Example-4/Report-word.docx' 'Example-4/Report-word.md'
#>
#> 16/01/2026 - 14:34:28
#> i Info Mean Model Workflow completed in 0.2 minThe output report for Example 4 is shown below.
#> file:////home/runner/work/OSPSuite.ReportingEngine/OSPSuite.ReportingEngine/vignettes/Example-4/Report.html screenshot completedReport

Example 5
In example 5, there are 2 simulation sets, each one includes one
output path. Simulation set A includes output path
Organism|A|Concentration in container and has observed
data. Simulation set B includes output path
Organism|B|Concentration in container and has no
observed data. The simulate task needs to be re-run also in
this case since the simulation results are saved in files that use the
simulation set names.
Code
# Get the pkml simulation file: "MiniModel1.pkml"
simulationFileA <- system.file("extdata", "MiniModel1.pkml",
package = "ospsuite.reportingengine"
)
# Get the pkml simulation file: "MiniModel2.pkml"
simulationFileB <- system.file("extdata", "MiniModel2.pkml",
package = "ospsuite.reportingengine"
)
# Get the observation file and its dictionary
dataSource <- DataSource$new(
dataFile = observationFile,
metaDataFile = dictionaryFile
)
# Define the input parameters
outputA <- Output$new(
path = "Organism|A|Concentration in container",
displayName = "Simulated concentration of A",
displayUnit = "µmol/ml",
dataSelection = "MDV==0",
dataDisplayName = "Observed concentration of A"
)
outputB <- Output$new(
path = "Organism|B|Concentration in container",
displayName = "Simulated concentration of B",
displayUnit = "µmol/ml"
)
setA <- SimulationSet$new(
simulationSetName = "A",
simulationFile = simulationFileA,
outputs = outputA,
dataSource = dataSource
)
#> Warning in readLines(file): incomplete final line found on
#> '/home/runner/work/_temp/Library/ospsuite.reportingengine/extdata/tpDictionary.csv'
setB <- SimulationSet$new(
simulationSetName = "B",
simulationFile = simulationFileB,
outputs = outputB
)
# Create the workflow instance
workflow5 <-
MeanModelWorkflow$new(
simulationSets = c(setA, setB),
workflowFolder = "Example-5"
)
#> 16/01/2026 - 14:34:30
#> i Info Reporting Engine Information:
#> Date: 16/01/2026 - 14:34:30
#> User Information:
#> Computer Name: runnervmmtnos
#> User: runner
#> Login: unknown
#> System is NOT validated
#> System versions:
#> R version: R version 4.5.2 (2025-10-31)
#> OSP Suite Package version: 12.4.1.9001
#> OSP Reporting Engine version: 2.4.0.9005
#> tlf version: 1.6.2.9001
# Set the workflow tasks to be run
workflow5$activateTasks(c("simulate", "plotTimeProfilesAndResiduals"))
# Run the workflow
workflow5$runWorkflow()
#> 16/01/2026 - 14:34:30
#> i Info Starting run of Mean Model Workflow
#> 16/01/2026 - 14:34:30
#> i Info Starting run of Simulation task
#> 16/01/2026 - 14:34:30
#> i Info Splitting simulations for parallel run: 2 simulations split into 1 subsets
#> 16/01/2026 - 14:34:30
#> i Info Starting run of subset simulations
#> | | | 0% | |======================================================================| 100%
#> 16/01/2026 - 14:34:30
#> i Info Simulation task completed in 0 min
#> 16/01/2026 - 14:34:30
#> i Info Starting run of Plot Time profiles and Residuals task
#> 16/01/2026 - 14:34:30
#> i Info Starting run of Plot Time profiles and Residuals task for A
#> 16/01/2026 - 14:34:31
#> ! Warning incomplete final line found on '/home/runner/work/_temp/Library/ospsuite.reportingengine/extdata/tpDictionary.csv'
#> 16/01/2026 - 14:34:39
#> i Info Starting run of Plot Time profiles and Residuals task for B
#> 16/01/2026 - 14:34:48
#> i Info Plot Time profiles and Residuals task completed in 0.3 min
#> 16/01/2026 - 14:34:48
#> i Info Executing: pandoc --embed-resources --standalone --wrap=none --toc --from=markdown+tex_math_dollars+superscript+subscript+raw_attribute --reference-doc="/home/runner/work/_temp/Library/ospsuite.reportingengine/extdata/reference.docx" --resource-path="Example-5" -t docx -o 'Example-5/Report-word.docx' 'Example-5/Report-word.md'
#>
#> 16/01/2026 - 14:34:48
#> i Info Mean Model Workflow completed in 0.3 minThe output report for Example 5 is shown below.
#> file:////home/runner/work/OSPSuite.ReportingEngine/OSPSuite.ReportingEngine/vignettes/Example-5/Report.html screenshot completedReport

Example 6
Example 6 shows a goodness of fit for a population workflow. In this example, there is no observed data, only one simulation set and one output are requested.
Code
# Get the pkml simulation file: "MiniModel2.pkml"
simulationFile <- system.file("extdata", "MiniModel2.pkml",
package = "ospsuite.reportingengine"
)
populationFile <- system.file("extdata", "Pop500_p1p2p3.csv",
package = "ospsuite.reportingengine"
)
# Define the input parameters
outputA <- Output$new(
path = "Organism|A|Concentration in container",
displayName = "Concentration of A",
displayUnit = "µmol/ml"
)
setA <- PopulationSimulationSet$new(
simulationSetName = "A",
simulationFile = simulationFile,
populationFile = populationFile,
outputs = outputA
)
# Create the workflow instance
workflow6 <-
PopulationWorkflow$new(
workflowType = PopulationWorkflowTypes$parallelComparison,
simulationSets = setA,
workflowFolder = "Example-6"
)
#> 16/01/2026 - 14:34:51
#> i Info Reporting Engine Information:
#> Date: 16/01/2026 - 14:34:51
#> User Information:
#> Computer Name: runnervmmtnos
#> User: runner
#> Login: unknown
#> System is NOT validated
#> System versions:
#> R version: R version 4.5.2 (2025-10-31)
#> OSP Suite Package version: 12.4.1.9001
#> OSP Reporting Engine version: 2.4.0.9005
#> tlf version: 1.6.2.9001
# Set the workflow tasks to be run
workflow6$activateTasks(c("simulate", "plotTimeProfilesAndResiduals"))
# Run the workflow
workflow6$runWorkflow()
#> 16/01/2026 - 14:34:51
#> i Info Starting run of Population Workflow for parallelComparison
#> 16/01/2026 - 14:34:51
#> i Info Starting run of Simulation task
#> 16/01/2026 - 14:34:51
#> i Info Starting run of Population Simulations
#> | | | 0%
#> 16/01/2026 - 14:34:51
#> i Info Starting run of A (Pop500_p1p2p3)
#> | |======================================================================| 100%
#>
#> 16/01/2026 - 14:34:51
#> i Info Simulation task completed in 0 min
#> 16/01/2026 - 14:34:51
#> i Info Starting run of Plot Time profiles and Residuals task
#> 16/01/2026 - 14:34:51
#> i Info Starting run of Plot Time profiles and Residuals task for A
#> 16/01/2026 - 14:34:54
#> i Info Plot Time profiles and Residuals task completed in 0 min
#> 16/01/2026 - 14:34:54
#> i Info Executing: pandoc --embed-resources --standalone --wrap=none --toc --from=markdown+tex_math_dollars+superscript+subscript+raw_attribute --reference-doc="/home/runner/work/_temp/Library/ospsuite.reportingengine/extdata/reference.docx" --resource-path="Example-6" -t docx -o 'Example-6/Report-word.docx' 'Example-6/Report-word.md'
#>
#> 16/01/2026 - 14:34:54
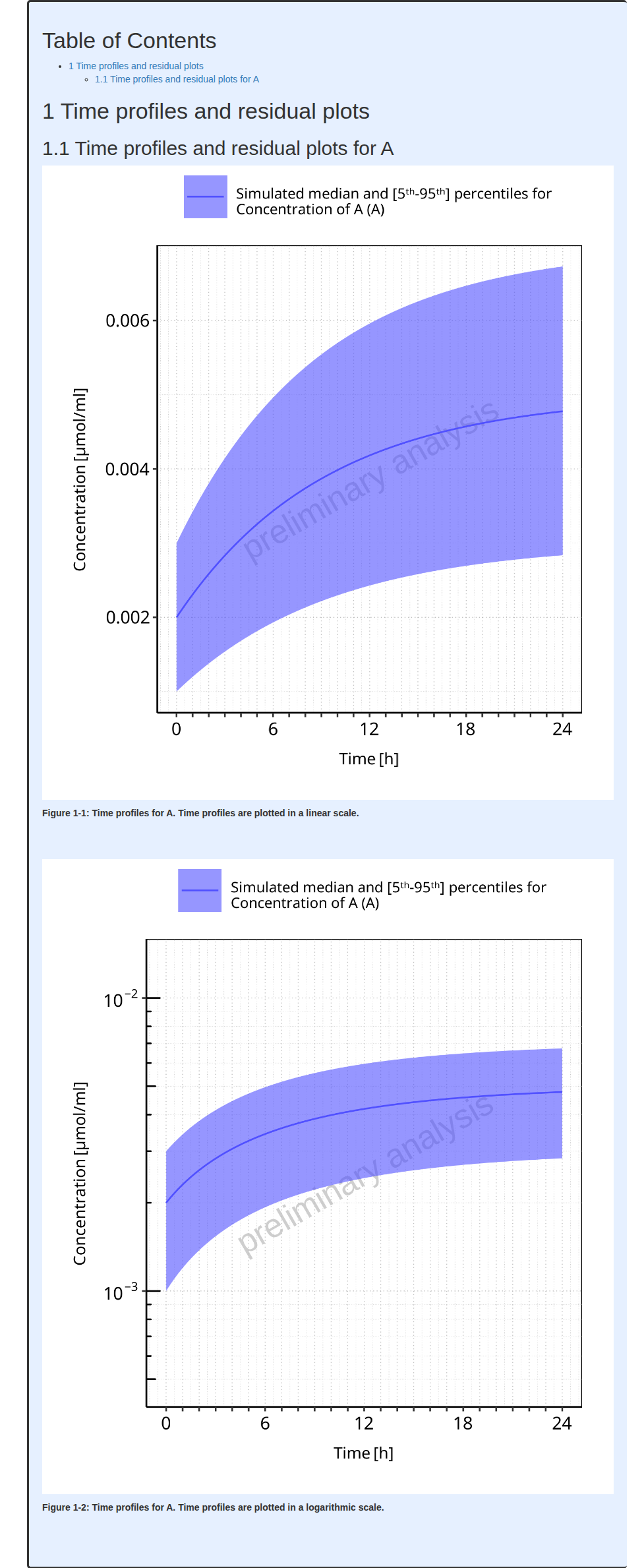
#> i Info Population Workflow for parallelComparison completed in 0.1 minThe output report for Example 6 is shown below. The 2 figures of the report (linear and log scales) include median, mean 5th and 95th percentiles of the population.
#> file:////home/runner/work/OSPSuite.ReportingEngine/OSPSuite.ReportingEngine/vignettes/Example-6/Report.html screenshot completedReport
Example 7
Example 7 add observed data to the previous workflow.
Code
# Get the pkml simulation file: "MiniModel2.pkml"
simulationFile <- system.file("extdata", "MiniModel2.pkml",
package = "ospsuite.reportingengine"
)
populationFile <- system.file("extdata", "Pop500_p1p2p3.csv",
package = "ospsuite.reportingengine"
)
# Get the observation file and its dictionary
dataSource <- DataSource$new(
dataFile = observationFile,
metaDataFile = dictionaryFile
)
# Define the input parameters
outputA <- Output$new(
path = "Organism|A|Concentration in container",
displayName = "Simulated concentration of A",
displayUnit = "µmol/ml",
dataSelection = "MDV==0",
dataDisplayName = "Observed concentration of A"
)
setA <- PopulationSimulationSet$new(
simulationSetName = "A",
simulationFile = simulationFile,
populationFile = populationFile,
outputs = outputA,
dataSource = dataSource
)
#> Warning in readLines(file): incomplete final line found on
#> '/home/runner/work/_temp/Library/ospsuite.reportingengine/extdata/tpDictionary.csv'
# Create the workflow instance
workflow7 <-
PopulationWorkflow$new(
workflowType = PopulationWorkflowTypes$parallelComparison,
simulationSets = setA,
workflowFolder = "Example-6"
)
#> 16/01/2026 - 14:34:55
#> i Info Reporting Engine Information:
#> Date: 16/01/2026 - 14:34:55
#> User Information:
#> Computer Name: runnervmmtnos
#> User: runner
#> Login: unknown
#> System is NOT validated
#> System versions:
#> R version: R version 4.5.2 (2025-10-31)
#> OSP Suite Package version: 12.4.1.9001
#> OSP Reporting Engine version: 2.4.0.9005
#> tlf version: 1.6.2.9001
# Set the workflow tasks to be run
workflow7$activateTasks(StandardPlotTasks$plotTimeProfilesAndResiduals)
workflow7$inactivateTasks(StandardSimulationTasks$simulate)
# Run the workflow
workflow7$runWorkflow()
#> 16/01/2026 - 14:34:55
#> i Info Starting run of Population Workflow for parallelComparison
#> 16/01/2026 - 14:34:55
#> i Info Starting run of Plot Time profiles and Residuals task
#> 16/01/2026 - 14:34:55
#> i Info Starting run of Plot Time profiles and Residuals task for A
#> 16/01/2026 - 14:34:56
#> ! Warning incomplete final line found on '/home/runner/work/_temp/Library/ospsuite.reportingengine/extdata/tpDictionary.csv'
#> 16/01/2026 - 14:35:05
#> i Info Plot Time profiles and Residuals task completed in 0.2 min
#> 16/01/2026 - 14:35:05
#> i Info Executing: pandoc --embed-resources --standalone --wrap=none --toc --from=markdown+tex_math_dollars+superscript+subscript+raw_attribute --reference-doc="/home/runner/work/_temp/Library/ospsuite.reportingengine/extdata/reference.docx" --resource-path="Example-6" -t docx -o 'Example-6/Report-word.docx' 'Example-6/Report-word.md'
#>
#> 16/01/2026 - 14:35:06
#> i Info Population Workflow for parallelComparison completed in 0.2 minThe output report for Example 7 is shown below. The residuals are calculated on the median simulated values.
#> file:////home/runner/work/OSPSuite.ReportingEngine/OSPSuite.ReportingEngine/vignettes/Example-6/Report.html screenshot completedReport