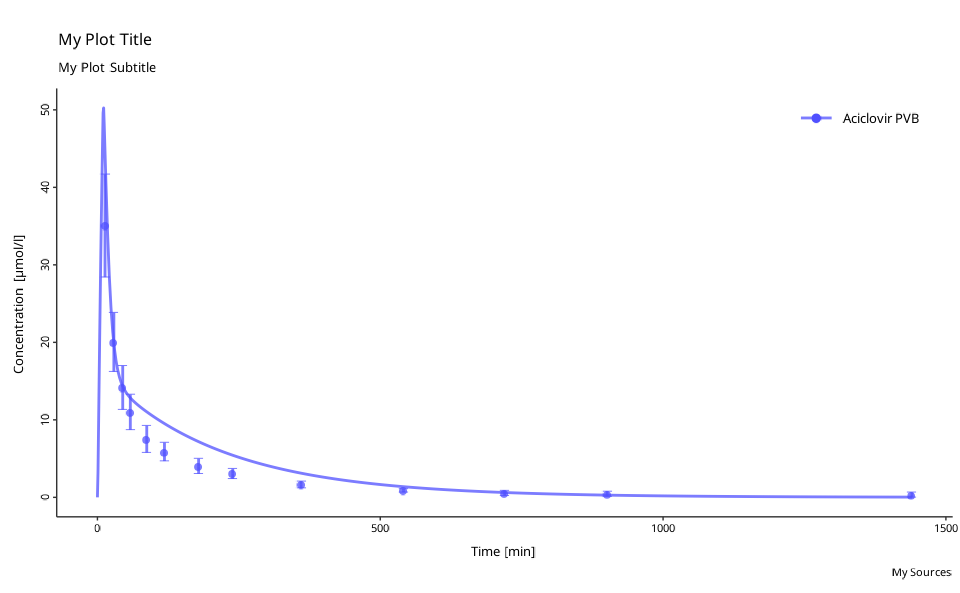
Time-profile plot of individual data
Source:R/plot-individual-time-profile.R
plotIndividualTimeProfile.RdTime-profile plot of individual data
Usage
plotIndividualTimeProfile(
dataCombined,
defaultPlotConfiguration = NULL,
showLegendPerDataset = FALSE
)Arguments
- dataCombined
A single instance of
DataCombinedclass containing both observed and simulated datasets to be compared.- defaultPlotConfiguration
A
DefaultPlotConfigurationobject, which is anR6class object that defines plot properties.- showLegendPerDataset
Logical flag to display separate legend entries for observed and simulated datasets, if available. This is experimental and may not work reliably when both observed and simulated datasets > 1. Defaults to
FALSE.
Examples
# simulated data
simFilePath <- system.file("extdata", "Aciclovir.pkml", package = "ospsuite")
sim <- loadSimulation(simFilePath)
simResults <- runSimulations(sim)[[1]]
outputPath <- "Organism|PeripheralVenousBlood|Aciclovir|Plasma (Peripheral Venous Blood)"
# observed data
obsData <- lapply(
c("ObsDataAciclovir_1.pkml", "ObsDataAciclovir_2.pkml", "ObsDataAciclovir_3.pkml"),
function(x) loadDataSetFromPKML(system.file("extdata", x, package = "ospsuite"))
)
names(obsData) <- lapply(obsData, function(x) x$name)
# Create a new instance of `DataCombined` class
myDataCombined <- DataCombined$new()
# Add simulated results
myDataCombined$addSimulationResults(
simulationResults = simResults,
quantitiesOrPaths = outputPath,
groups = "Aciclovir PVB"
)
# Add observed data set
myDataCombined$addDataSets(obsData$`Vergin 1995.Iv`, groups = "Aciclovir PVB")
# Create a new instance of `DefaultPlotConfiguration` class
myPlotConfiguration <- DefaultPlotConfiguration$new()
myPlotConfiguration$title <- "My Plot Title"
myPlotConfiguration$subtitle <- "My Plot Subtitle"
myPlotConfiguration$caption <- "My Sources"
# plot
plotIndividualTimeProfile(myDataCombined, myPlotConfiguration)